March 2020, By Amir Hossein Namadchi
This is an OpenSeesPy simulation of a simple linear 3 DOF system introduced by Bathe and Noh. The model was used to demonstrate the ability of Bathe method in filtering out the unwanted artificial high-frequency responses. The effectiveness of numerical dissipation in a specific time integration algorithm could be evaluated by analyzing this benchmark model problem [1].

import numpy as np
import opensees.openseespy as ops
import matplotlib.pyplot as plt
No units were defined in the original paper. So, I just assume the base units as follows:
## Units
m = 1 # Meters
KN = 1 # KiloNewtons
s = 1 # Seconds
kg = KN*(s**2)/m # mass unit (derived)
Model specifications are defined as follows:
# Nodal mass values
m_1, m_2, m_3 = 0*kg, 1*kg, 1*kg
# Modulus of Elasticity
E_1, E_2 = (10.0**7)*(KN/m**2) , 1.0*(KN/m**2)
# Prescribed displacment @ node 1
d_p = lambda t: np.sin(1.2*t)*m
# Dynamic Analysis Parameters
dt = 0.2618
time = 10
Here’s the exact solution for comparison. It can be obtained using modal decomposition technique, considering only the lowest frequency plus the static correction [1].
d_2_ex = lambda t : (2.7273e-7)*np.sin(t) + np.sin(1.2*t)
d_3_ex = lambda t : (2.7273)*np.sin(t) - (2.2727)*np.sin(1.2*t)
v_2_ex = lambda t : (2.7273e-7)*np.cos(t) + 1.2*np.cos(1.2*t)
v_3_ex = lambda t : (2.7273)*np.cos(t) - 1.2*(2.2727)*np.cos(1.2*t)
a_2_ex = lambda t : -(2.7273e-7)*np.sin(t) - 1.2*1.2*np.sin(1.2*t)
a_3_ex = lambda t : -(2.7273)*np.sin(t) + 1.2*1.2*(2.2727)*np.sin(1.2*t)
Model Construction
Use ZeroLength elements to model the spings. Note that nodal masses are added via mass option of the node command.
def build_model():
model = ops.Model(ndm=1, ndf=1)
# Adding nodes
for n, mass in enumerate([m_1,m_2,m_3]):
model.node(n+1, 0.0, mass=mass)
# Defining Material (spring stiffness)
model.uniaxialMaterial('Elastic', 1, E_1)
model.uniaxialMaterial('Elastic', 2, E_2)
# Add springs
model.element('ZeroLength', 1, *[1,2], '-mat', 1, '-dir', 1)
model.element('ZeroLength', 2, *[2,3], '-mat', 2, '-dir', 1)
print('Model built successfully!')
return model
Dynamic Analysis
To use different time integration algorithms, the function do_dynamic_analysis is defined herein. Just pass the time, dt and integrator_params and run it multiple times with different integrators. Here, Newmark with $\gamma=0.5$ and $\beta=0.25$ (a non-dissipative algorithm) and the Bathe method, named as 'TRBDF2' (with asymptotic annihilation property) are used.
def do_dynamic_analysis(time, dt, integrator_params):
model = build_model()
time_domain = np.arange(0,time,dt)
# Constraints
model.timeSeries('Path', 1,
dt=dt,
values=np.vectorize(d_p)(time_domain),
time=time_domain)
model.pattern('Plain', 1, 1)
model.sp(1, 1, 1)
# Analysis
model.constraints('Transformation')
model.numberer('Plain')
model.system('ProfileSPD')
model.algorithm('Linear')
model.integrator(*integrator_params)
model.analysis('Transient')
time_lst =[0] # list to hold time stations for plotting
res_node_2 = [[0,0,0]] # response params of node 2
res_node_3 = [[0,0,0]] # response params of node 3
for i in range(len(time_domain)):
model.analyze(1, dt)
time_lst.append(model.getTime())
res_node_2.append([model.nodeDisp(2,1),
model.nodeVel(2,1),
model.nodeAccel(2,1)])
res_node_3.append([model.nodeDisp(3,1),
model.nodeVel(3,1),
model.nodeAccel(3,1)])
print('Done with ', integrator_params[0],'!')
return {'time_list':np.array(time_lst),
'Node 2': np.array(res_node_2),
'Node 3': np.array(res_node_3)}
Let’s perform analysis:
newmark = do_dynamic_analysis(time, dt,['Newmark',0.5,0.25])
bathe = do_dynamic_analysis(time, dt,['TRBDF2'])
Output:
Model built successfully!
Done with Newmark !
Model built successfully!
Done with TRBDF2 !
Visualization
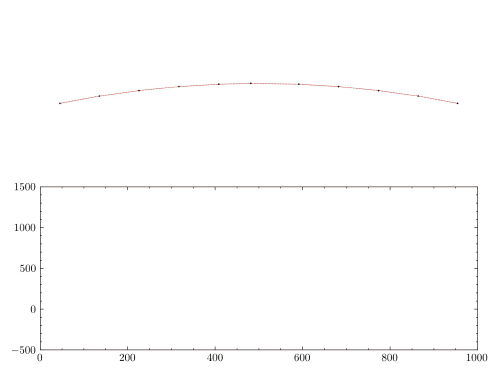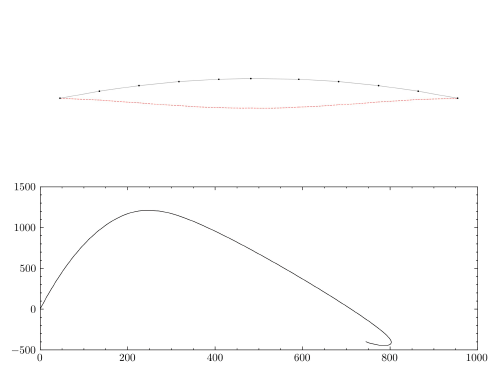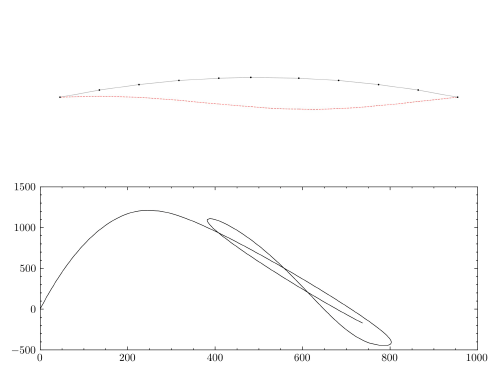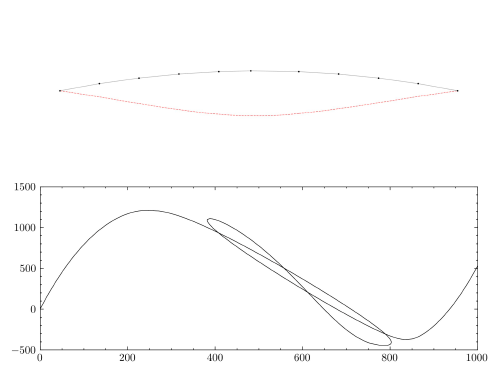
plt.figure(figsize=(12,4))
plt.plot(np.arange(0, time, 0.2*dt),
np.vectorize(v_2_ex)(np.arange(0,time,0.2*dt)),
color = '#7D3C98', linewidth = 2, label = 'Exact')
plt.plot(newmark['time_list'], newmark['Node 2'][:,1],
color = '#d62d20', linewidth=1.5, ls='--', label = 'Newmark (CAAM)' )
plt.plot(bathe['time_list'], bathe['Node 2'][:,1],
color = '#008000', linewidth=1.75, label = 'Bathe (TRBDF2)', marker = 'o',ls=':')
plt.ylabel('Velocity of Node 2 (m/s)', {'fontstyle':'italic','size':14})
plt.xlabel('Time (sec)', {'fontstyle':'italic','size':14})
plt.xlim([0.0, time])
plt.ylim([-3, 3])
plt.grid()
plt.legend()
plt.yticks(fontsize = 14)
plt.xticks(fontsize = 14);
Output:
<Figure size 1200x400 with 1 Axes>
plt.figure(figsize=(12,4))
plt.plot(np.arange(0, time, 0.2*dt),
np.vectorize(a_2_ex)(np.arange(0,time,0.2*dt)),
color = '#7D3C98', linewidth = 2, label = 'Exact')
plt.plot(newmark['time_list'], newmark['Node 2'][:,2],
color = '#d62d20', linewidth=1.5, ls='--', label = 'Newmark (CAAM)' )
plt.plot(bathe['time_list'], bathe['Node 2'][:,2],
color = '#008000', linewidth=1.75, label = 'Bathe (TRBDF2)', marker = 'o',ls=':')
plt.ylabel('Acceleration of Node 2 m/s^2', {'fontstyle':'italic','size':14})
plt.xlabel('Time (sec)', {'fontstyle':'italic','size':14})
plt.xlim([0.0, time])
#plt.ylim([-30, 30])
plt.grid()
plt.legend()
plt.yticks(fontsize = 14)
plt.xticks(fontsize = 14);
Output:
<Figure size 1200x400 with 1 Axes>
Closure
As expected, non-dissipative methods like CAAM are unable to represent the exact response due to spurious high frequency oscillations. On the other hand, TRBDF2 scheme can effectively damp out these oscillations via algorithmic damping.
References
-
Bathe, K.J. and Noh, G., 2012. Insight into an implicit time integration scheme for structural dynamics. Computers & Structures, 98, pp.1-6.
-
Namadchi, A.H., Jandaghi, E. and Alamatian, J., 2020. A new model-dependent time integration scheme with effective numerical damping for dynamic analysis. Engineering with Computers, pp.1-16.